Standalone C++ software testing, debugging and analysis with python packages: NumPy, matplotlib, ...
Standalone C++ software testing, debugging and analysis
with python packages: NumPy, matplotlib, pyvista, ...
- (p1) Three Laws of Good Software Design + Development
- (p2) Power of Serialization + De-Serialization
- (p3-12) Opticks : Example Open Source Package
- (p13-14) Best IDE to use : Unix Command Line + bash shell
- (p15-16) Anaconda Distribution
- (p17-22) NumPy : Fundamental to Python Data Ecosystem
- (p23) NPY minimal file format : metadata header + data buffer : trivial to parse
- (p24-27) Matplotlib, PyVista
- (p28-31) Standalone Software Test Example : U4Mesh_test.sh
- (p32-36) stamp_test.sh : remote/local workflow example
- (p37-43) Standalone Test Example : mandelbrot.sh, mandelbrot.cc
- (p44) Bash function Tip
- (p45) IPython launch customization
- (p46-87) Python Data Science Handbook
- (p89-90) Concluding
Simon C Blyth, IHEP, CAS — Hohhot Meeting — 30 Sept 2023
Three Laws of Good Software Design + Development
Curse of Software Dependencies
- slow dev. cycle : ~mins build + run
- only "high" level tests of whole packages
- => LOTS OF (OLD) BUGS
Solution : Divide and Conquer => split into units
- fast dev. cycle : change unit + rerun in < 1s
- fine grained test => EASY FIND/FIX BUGS
HOW CAN SOFTWARE BE SPLIT APART:
- design to minimize dependencies of "unit"
- less dependencies => more useful
- Literally, as usable in more situations
- persist objects with serialization
- save to file, load from file
- enables split dependencies + processing
Serialization : sw flexibility superpower
|
|
|
Power of Serialization + De-Serialization
What is Serialization ?
Convert objects into stream of bytes, enabling:
- write to file
- transfer over network
- upload to GPU address space
Handle data with standard tools
Favor simple data structures
- arrays of simple struct
- simplify serialization de-serialization
Power arises from dependency "suspension":
- many deps to create objects, much fewer to use
- => Huge potential for software simplification
Under-used : as objects often over designed
- design for easy serialize/de-serialize
- => flexible data handling
Opticks : GPU Optical Photon Simulation Package
Opticks bitbucket
Opticks : Open source project
- based on NVIDIA OptiX : GPU ray tracing
- many thousands of classes, structs
- thousands of unit tests
- accelerates optical photon simulation
- speedup factor > 1500x
Importance of Unit tests
- not just for testing
- ideal way to recall functionality
- integral part of development
- run tests as add new features
https://bitbucket.org/simoncblyth/opticks/
Optical Photon Simulation Problem...
Huge CPU Memory+Time Expense
- JUNO Muon Simulation Bottleneck
- ~99% CPU time, memory constraints
- Ray-Geometry intersection Dominates
- simulation is not alone in this problem...
- Optical photons : naturally parallel, simple :
- produced by Cherenkov+Scintillation
- yield only Photomultiplier hits
Optical Photon Simulation ≈ Ray Traced Image Rendering
Not a Photo, a Calculation
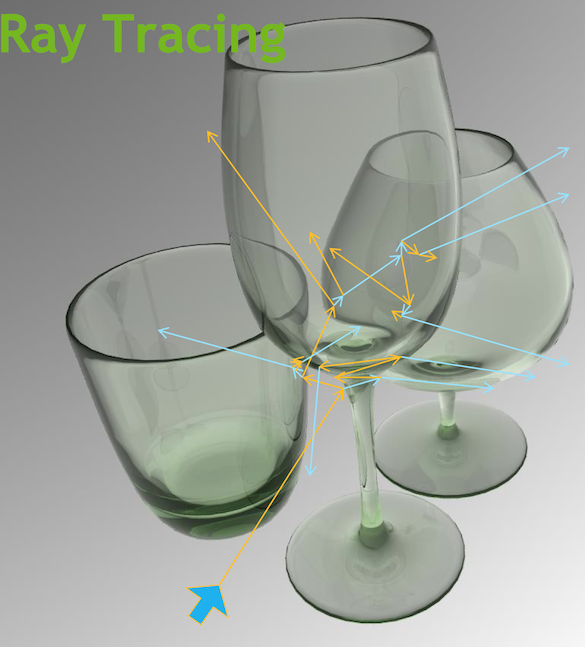
- simulation
- photon parameters at sensors (PMTs)
- rendering
- pixel values at image plane
Much in common : geometry, light sources, optical physics
- both limited by ray geometry intersection, aka ray tracing
Many Applications of ray tracing :
- advertising, design, architecture, films, games,...
- -> huge efforts to improve hw+sw over 30 yrs
Geant4 + Opticks + NVIDIA OptiX 7 : Hybrid Workflow
| https://bitbucket.org/simoncblyth/opticks |
Opticks API : split according to dependency -- Optical photons are GPU "resident", only hits need to be copied to CPU memory
[9]cxr_i0_t8,_-1 : EXCLUDE SLOWEST
JUNO Opticks OptiX 7 Ray-trace
CSGFoundry CPU/GPU Geometry
- purely analytic CSG, no triangles
cxr_min__eye_-10,0,0__zoom_0.5__tmin_0.1__sChimneyAcrylic_increased_TMAX.jpg
ELV=t94,95 ./cxr_min.sh ## skip sTarget sAcrylic
cxr_min__eye_-10,0,-30__zoom_0.5__tmin_0.1__sChimneyAcrylic_photon_eye_view.jpg
ELV=t94,95 ./cxr_min.sh ## skip sTarget sAcrylic : upwards view
scan-pf-check-GUI-TO-SC-BT5-SD
scan-pf-check-GUI-TO-BT5-SD
Large packages like Opticks are built test-by-test
Standalone Test
- runs without many dependencies
- sometime dependencies are "mocked"
Complex functionality built from thousands of simple units
- each unit is simple
- each unit has its own simple test
- where possible tests are designed to be standalone
Unit tests:
- written at the same time as the unit
- kept in a "tests" directory next to the unit
- updated as functionality is added to the unit
Act of writing test while adding functionality
- improves API : as developer sees user perspective
Best IDE to use : Unix Command Line + bash shell
What is wrong with IDEs like "Visual Studio Code"?
- hides commands and errors
- like typing while wearing boxing gloves
- pain to describe setup : this window, this box,...
- megabytes of bloat-ware : slow
- big project files, vendor lock in
- often not the target build system
- prevents learning tools directly
Suggest: Avoid IDEs when learning
- Just adds extra complex layer
- Imperfect interface ontop of tools
Using tools directly : you will learn faster
- bash shell
- coordinate an infinity of tools
- vim (or other minimal editor)
- text editor
- compiler (gcc/clang etc..)
- build code
- ipython (interactive python shell)
- analysis and debugging
- rsync
- transfer files between machines
- git,zip,curl,...
- plethora of tools : whatever you need
Advantages of NOT using an IDE
- Standard tools : that work almost anywhere
- Easy to automate : just write bash scripts
- Nothing hidden : commands and errors directly
- Freedom to use(or create) the tool appropriate to the task
- Arrange same environment on remote server anywhere
- Work with Open source projects : Cross-Platform
Best_IDE_bash_command_line.png
Anaconda : Distribution of ~500 Data Science Pkgs
MIXING ENVIRONMENTS CAUSES BREAKAGES
What will happen ? Mysterious bugs when trying to run/build packages in all environments.
| BEST PRACTICE |
|
Example bash function that enables anaconda environment:
bes3_conda ()
{
type $FUNCNAME;
eval "$(/cvmfs/bes3.ihep.ac.cn/bes3sw/ExternalLib/contrib/anaconda/2023.07/bin/conda shell.bash hook)"
}
- THAT WILL CLASH WITH BES3 ENVIRONMENT
Anaconda Distribution : Highlight packages
Foundation Packages (This presentation focusses on Foundation Packages)
- numpy : core array
- ipython : enhanced interactive python shell
- matplotlib : plotting
Details
- scipy : optimization, integration, interpolation, ...
- sympy : symbolic algebraic manipulations
- pandas : data analysis and manipulation tool
- scikit-image : python image processing
- scikit-learn : python machine learning
Utilities
- pillow : Python Imaging Library
- lxml : python XML handling binding to C libraries libxml2 libxslt
- zeromq : message queue system
- sphinx : RST based documentation system
NumPy : Fundamental to Python Data Ecosystem
- central nature of NumPy to the Python world makes mastering it exceedingly useful
- understanding it, enables C/C++ performance with python brevity + ease of development
https://docs.scipy.org/doc/numpy/user/quickstart.html
http://www.scipy-lectures.org/intro/index.html
https://github.com/donnemartin/data-science-ipython-notebooks
NumPy_Logo.png
https://numpy.org/
NumPy_Broad.png
The Zen of Numpy, by its creator, Travis Oliphant
Explanation
- C++ new scatters memory allocation
- Arrays use contiguous blocks of memory
- C++ std::vector uses contiguous memory (but reallocs as it grows)
Allocated memory layout is critical
Strided is better than scattered. Contiguous is better than strided. Descriptive is better than imperative[1] (e.g. data-types). Array-orientated is better than object-oriented. Broadcasting is a great idea -- use where possible! Vectorized is better than an explicit loop. Unless its complicated -- then use Cython or numexpr. Think in higher dimensions.
My take : best tool depends on nature of data
- NumPy shines for large[2] and simple data ; splitting data to make it simple brings other benefits !
- NumPy holistic approach : prepares you for vectorized and parallel processing
- no-looping makes for an terse, intuitive interactive interface
[1] imperative means step by step how to do something
[2] but not so large that has trouble fitting in memory, np.memmap is possible but better to avoid for simplicity
NumPy_views
NumPy views : same data, diff. view
b is a view of a with different shape
NumPy : Python flexibility+brevity at C speed
github.com/simoncblyth/np
- inspired by NumPy : does not depend on it
- implements NumPy serialization file format, into which almost everything(*) is persisted
- Opticks analysis/debugging done with NumPy and IPython
(*) gensteps, photons, hits, analytic CSG geometry shapes, transforms, triangulated geometry vertices, triangles, material/surface properties ...
“fundamental package for scientific computing with Python”
NumPy arrays : simply an interface to C memory buffers
- extreme simplicity : simple interop with C, C++, CUDA
- slices are no-copy "views" of underlying buffers
- reshape/transpose : just changes metadata
- fast memcpy, cudaMemcpy, serialization/deserialization
Very terse, no-loop python interface to C performance
- array-oriented computing (C loops under python control)
- interactive handling of very large N-dimensional arrays
- easily manipulate million item arrays from python
- faster + more convenient than dealing with millions of C++ objects
“The NumPy array: a structure for efficient numerical computation”
NPY file format specification
https://github.com/numpy/numpy/blob/master/doc/neps/nep-0001-npy-format.rst
NPY minimal file format : metadata header + data buffer : trivial to parse
- data accessible from anywhere : C/C++/CUDA/python/... ; Simple to memcpy() or cudaMemcpy() to GPU
In [1]: a = np.arange(10) # array of 10 ints : 64 bit, 8 bytes each
In [2]: np.save("a.npy", a ) # persist the array : serializing it into a file
In [3]: a2 = np.load("a.npy") # load array from file into memory
In [4]: assert np.all( a == a2 ) # check all elements the same
In [5]: !xxd a.npy # run xxd in shell to hexdump the byte contents of the file
00000000: 934e 554d 5059 0100 7600 7b27 6465 7363 .NUMPY..v.{ desc
00000010: 7227 3a20 273c 6938 272c 2027 666f 7274 r': '<i8', 'fort
00000020: 7261 6e5f 6f72 6465 7227 3a20 4661 6c73 ran_order : Fals
00000030: 652c 2027 7368 6170 6527 3a20 2831 302c e, 'shape': (10, # minimal metadata : type, shape
00000040: 292c 207d 2020 2020 2020 2020 2020 2020 ), }
00000050: 2020 2020 2020 2020 2020 2020 2020 2020
00000060: 2020 2020 2020 2020 2020 2020 2020 2020
00000070: 2020 2020 2020 2020 2020 2020 2020 200a . # 128 bytes of header
00000080: 0000 0000 0000 0000 0100 0000 0000 0000 ................
00000090: 0200 0000 0000 0000 0300 0000 0000 0000 ................
000000a0: 0400 0000 0000 0000 0500 0000 0000 0000 ................ # data buffer
000000b0: 0600 0000 0000 0000 0700 0000 0000 0000 ................
000000c0: 0800 0000 0000 0000 0900 0000 0000 0000 ................
In [6]: !ls -l a.npy # small 128 byte header + (8 bytes per integer)*10 = 208 bytes total
-rw-r--r-- 1 blyth staff 208 Sep 13 11:01 a.npy
In [7]: a
Out[7]: array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
In [8]: a.shape
Out[8]: (10,)
One possibility for compression with blosc http://bcolz.blosc.org/en/latest/intro.html
Matplotlib_Logo.png
PyVista_Logo.png
https://docs.pyvista.org/version/stable/
ntds3_ana
~/j/ntds/ntds3.sh info_ana
ntds3_ana 2
PyVista : 3D Plotting
- GPU accelerated : handles millions of data points
- Much better than Matplotlib for 3D
Standalone Software Test Example : U4Mesh_test.sh
What is a Standalone Test ?
- Very Simple => Very Fast
- tests small "unit" of software
- U4Mesh.h
- => simple fast build (no Makefile,..)
- Serialization : C++ -> .npy -> NumPy
- => analyse in python
- Communicate bash/C++/py : envvar FOLD
<= eg: Standalone Bash Script
./U4Mesh_test.sh info ./U4Mesh_test.sh build ./U4Mesh_test.sh run ./U4Mesh_test.sh ana
https://bitbucket.org/simoncblyth/opticks/src/master/u4/tests/U4Mesh_test.sh
#!/bin/bash -l
cd $(dirname $BASH_SOURCE)
name=U4Mesh_test
export FOLD=/tmp/$name
bin=$FOLD/$name
mkdir -p $FOLD
vars="BASH_SOURCE name FOLD bin"
defarg="info_build_run_ana"
arg=${1:-$defarg}
g4- ; clhep-
if [ "${arg/info}" != "$arg" ]; then
for var in $vars ; do
printf "%20s : %s \n" "$var" "${!var}" ; done
fi
if [ "${arg/build}" != "$arg" ]; then
gcc $name.cc
-g -std=c++11 -lstdc++ -I.. -I$HOME/np
-I$(clhep-prefix)/include -L$(clhep-prefix)/lib
-I$(g4-prefix)/include/Geant4 -L$(g4-prefix)/lib
-lG4global -lG4geometry -lG4graphics_reps -lCLHEP
-o $bin
[ $? -ne 0 ] && echo $BASH_SOURCE build error && exit 1
fi
if [ "${arg/run}" != "$arg" ]; then
$bin
[ $? -ne 0 ] && echo $BASH_SOURCE run error && exit 2
fi
if [ "${arg/ana}" != "$arg" ]; then
${IPYTHON:-ipython} --pdb -i $name.py
[ $? -ne 0 ] && echo $BASH_SOURCE ana error && exit 3
fi
exit 0
|
Using NP.hh NPFold.h Serialization from U4Mesh.h
Serialize + Save
https://bitbucket.org/simoncblyth/opticks/src/master/u4/U4Mesh.h
inline NPFold* U4Mesh::serialize() const
{
NPFold* fold = new NPFold ;
fold->add("vtx", vtx );
fold->add("fpd", fpd );
return fold ;
}
inline void U4Mesh::save(const char* base) const
{
NPFold* fold = serialize();
fold->save(base);
}
|
- NP.hh
- NumPy .npy file format serialization
- NPFold.h
- In memory folders-of-folders of NP.hh arrays
https://github.com/simoncblyth/np
#include "G4Polyhedron.hh" #include "NP.hh" #include "NPFold.h" struct U4Mesh { const G4VSolid* solid ; G4Polyhedron* poly ; int nv,nf ; NP* vtx ; double* vtx_ ; NP* fpd ; ... }; inline U4Mesh::U4Mesh(const G4VSolid* solid_): solid(solid_), poly(solid->CreatePolyhedron()), nv(poly->GetNoVertices()), nf(poly->GetNoFacets()), vtx(NP::Make<double>(nv, 3)), vtx_(vtx->values<double>()), fpd(nullptr) { for(int i=0 ; i < nv ; i++){ G4Point3D point = poly->GetVertex(i+1) ; vtx_[3*i+0] = point.x() ; vtx_[3*i+1] = point.y() ; vtx_[3*i+2] = point.z() ; } ... } |
Loading Serialized U4Mesh with NumPy, Visualize with PyVista
PyVista 3D Visualization
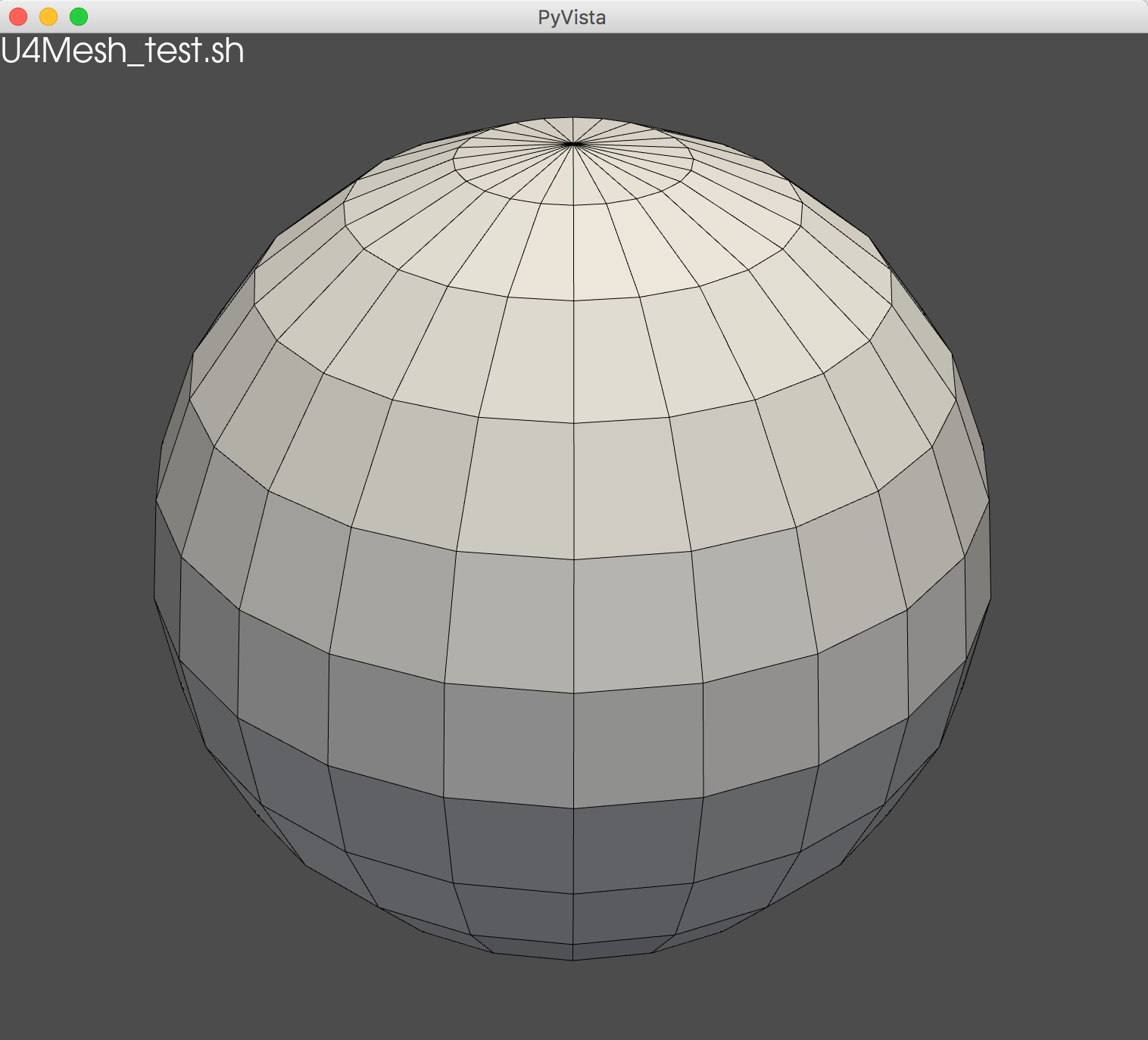
./U4Mesh_test.sh ana
u4/tests/U4Mesh_test.cc
#include "G4Orb.hh"
#include "U4Mesh.h"
int main()
{
G4Orb* solid = new G4Orb("Orb", 100);
U4Mesh::Save(solid, "$FOLD");
return 0;
};
|
u4/tests/U4Mesh_test.py
#!/usr/bin/env python
from np.fold import Fold
import numpy as np
import pyvista as pv
SIZE = np.array([1280, 720])
if __name__ == '__main__':
f = Fold.Load("$FOLD",symbol="f")
print(repr(f))
pd = pv.PolyData(f.vtx, f.fpd) # tri and/or quad
pl = pv.Plotter(window_size=SIZE*2)
pl.add_text("U4Mesh_test.sh", position="upper_left")
pl.add_mesh(pd, show_edges=True, lighting=True )
pl.show()
pass
|
U4Mesh_test2_xjfcSolid_review.png
GEOM=xjfcSolid ~/opticks/u4/tests/U4Mesh_test2.sh ana
stamp_test.cc example
Check time stamp overhead
- allocate NP array for 25M uint64_t
- collect 25M time stamps
- persist the stamps to .npy file
#include "stamp.h"
#include "NP.hh"
void test_performance()
{
static const int N = 1000000*25 ;
NP* t = NP::Make<uint64_t>(N) ;
uint64_t* tt = t->values<uint64_t>();
for(int i=0 ; i < N ; i++) tt[i] = stamp::Now();
t->save("$FOLD/tt.npy");
}
int main()
{
test_performance();
return 0 ;
}
stamp.h
C++ Timestamp Header
- convert std::chrono:::time_point into uint64_t
#pragma once
#include <chrono>
#include <string>
#include <sstream>
#include <iomanip>
struct stamp
{
static uint64_t Now();
static std::string Format(uint64_t t=0, const char* fmt="%FT%T.");
};
inline uint64_t stamp::Now()
{
using Clock = std::chrono::system_clock;
using Unit = std::chrono::microseconds ;
std::chrono::time_point<Clock> t0 = Clock::now();
return std::chrono::duration_cast<Unit>(t0.time_since_epoch()).count() ;
}
...
stamp_test.sh : Demonstrates Local/Remote Workflow
./stamp_test.sh info ## list bash variable values ./stamp_test.sh build ## compile C++ test into executable ./stamp_test.sh run ## run the executable ./stamp_test.sh grab ## rsync FOLD files from remote to local ./stamp_test.sh ana ## python analysis, plotting ./stamp_test.sh ls ## list files in FOLD
Simply by having this same code cloned from a git repo on local and remote machines : you can adopt a simple Local/Remote workflow.
The script automates what you could laboriously do in a manual way.
STAMP_ProcessHits_56us_Hama_0_0.png
STAMP_TT=257400,600 STAMP_ANNO=1 ~/j/ntds/stamp.sh ## inpho => Hama:0:0
(s0) beginPhoton (s1) finalPhoton ( ) pointPhoton (h0) ProcessHits[ (i0) ProcessHits] -- ProcessHits taking ~56us for SD
Exercise 0 : Clone, build, run time stamp example
Remote/Local
Common workflow : combining remote + local
- Clone the stamp repo onto both:
- remote server eg lxslc707.ihep.ac.cn
- local laptop/workstation
## clone from public repos on github.com (using git+https) git clone https://github.com/simoncblyth/stamp git clone https://github.com/simoncblyth/np ## OR from internal repos on code.ihep.ac.cn (using git+ssh) git clone git@code.ihep.ac.cn:blyth/stamp.git git clone git@code.ihep.ac.cn:blyth/np.git
- Build and run the test on remote server
- Grab timestamps from remote server onto local laptop
- Load timestamps into IPython on local laptop and examine the values
Standalone Test Example : mandelbrot.sh
Very Similar to U4Mesh_test.sh
- Fast cycle : changing code, param
- C++ -> .npy -> IPython
- include metadata for labelling
- envvar communication across C++/bash/py
<= eg: Standalone Bash Script
./mandelbrot.sh build ./mandelbrot.sh run ./mandelbrot.sh ana
https://github.com/simoncblyth/mandelbrot/blob/master/mandelbrot.sh
#!/bin/bash -l
usage(){ cat << EOU
mandelbrot.sh
===============
FOCUS=-1.45,0,0.05 ~/mandelbrot/mandelbrot.sh
FOCUS=-1.45,0,0.05 MIT=80 ~/mandelbrot/mandelbrot.sh
FOCUS=-1.45,0,0.05 MIT=50 ~/mandelbrot/mandelbrot.sh
EOU
}
cd $(dirname $BASH_SOURCE)
name=mandelbrot
defarg="build_run_ana"
arg=${1:-$defarg}
export FOLD=/tmp/$name
mkdir -p $FOLD
bin=$FOLD/$name
if [ "${arg/build}" != "$arg" ]; then
gcc $name.cc -I$HOME/np -std=c++11 -lstdc++ -o $bin
[ $? -ne 0 ] && echo $BASH_SOURCE build error && exit 1
fi
if [ "${arg/run}" != "$arg" ]; then
$bin
[ $? -ne 0 ] && echo $BASH_SOURCE run error && exit 2
fi
if [ "${arg/ana}" != "$arg" ]; then
${IPYTHON:-ipython} --pdb -i $name.py
[ $? -ne 0 ] && echo $BASH_SOURCE ana error && exit 1
fi
exit 0
|
Standalone Test Example : mandelbrot.cc
Continued : C++ Mandelbrot calc
...
std::complex<double> c0(X[0],Y[0]) ;
for(int iy=0 ; iy<NY ;iy++)
for(int ix=0 ; ix<NX ;ix++)
{
std::complex<double> c(ix*X[2], iy*Y[2]);
std::complex<double> z(0.0, 0.0);
int count(0) ;
while(std::norm(z)<MZZ && ++count < MIT-1)
{
z=z*z + c0 + c;
}
aa[iy*NX+ix] = std::min(count,MIT) ;
}
}
int main()
{
Mandelbrot m ;
m.a->save("$FOLD/a.npy");
return 0 ;
}
|
- C++ => Python/matplotlib : Presentation
- Connected with NP.hh serialization
#include <complex>
#include <array>
#include <vector>
#include "NP.hh"
struct Mandelbrot
{
const int MIT, NX, NY ;
const double MZZ, aspect ;
std::vector<double> F ;
std::array<double,3> X ;
std::array<double,3> Y ;
NP* a ;
unsigned char* aa ;
Mandelbrot();
};
inline Mandelbrot::Mandelbrot()
:
MIT(U::GetE<int>("MIT",255)),
NX(1280), NY(720),
MZZ(U::GetE<double>("MZZ",4.0)),
aspect(double(NX)/double(NY)),
F(*U::GetEnvVec<double>("FOCUS","-0.7,0,0.84375")),
a(NP::Make<unsigned char>(NY,NX)),
aa(a->values<unsigned char>())
{
X[0] = F[0] - F[2]*aspect ;
X[1] = F[0] + F[2]*aspect ;
X[2] = 2.*F[2]*aspect/double(NX) ;
Y[0] = F[1] - F[2] ;
Y[1] = F[1] + F[2] ;
Y[2] = 2.*F[2]/double(NY) ;
...
|
mandelbrot001.png
~/mandelbrot/mandelbrot.sh
mandelbrot000.png
FOCUS=-1.45,0,0.05 MIT=50 ~/mandelbrot/mandelbrot.sh
Standalone Test Example : mandelbrot.py
read_npy : standard file format
- NumPy : read a.npy : standard file format
- read metadata "sidecar" a_meta.txt
main : matplotlib/imshow
- also present metadata label
#!/usr/bin/env python import numpy as np SIZE = np.array([1280, 720]) import matplotlib.pyplot as plt def read_npy(path, d): path = os.path.expandvars(path) a = np.load(path) if not d is None: txtpath = path.replace(".npy","_meta.txt") lines = open(txtpath).read().splitlines() for line in lines: key, val = line.split(":") d[key] = val pass pass return a |
if __name__ == '__main__':
d = dict()
a = read_npy("$FOLD/a.npy", d)
d["CMAP"] = os.environ.get("CMAP", "prism")
cmap = getattr(plt.cm, d["CMAP"], None)
d["extent"] = list(map(float,(d["xmin"], d["xmax"], d["ymin"], d["ymax"] )))
label = "mandelbrot.sh : CMAP %(CMAP)s FOCUS %(FOCUS)s MZZ %(MZZ)s"
label += " MIT %(MIT)s extent %(extent)s "
fig, ax = plt.subplots(figsize=SIZE/100.)
fig.suptitle(label % d)
ax.imshow(a, extent=d["extent"], cmap=cmap)
fig.show()
|
Exercise 1 : Clone, build and run the Mandelbrot example
Assuming you already have : bash, git, python, ipython, matplotlib
- python related packages can conveniently be installed using conda
clone mandelbrot and np repos:
cd git clone https://github.com/simoncblyth/mandelbrot git clone https://github.com/simoncblyth/np
read the code downloaded, ask about things you do not understand
build and run:
cd ~/mandelbrot ./mandelbrot.sh
read the mandelbrot README and try FOCUS and max iterations MIT parameters as suggested
try changing the code : change the formula
Exercise 1 : Alternative URLs
If github is blocked for you:
cd git clone git@code.ihep.ac.cn:blyth/mandelbrot.git git clone git@code.ihep.ac.cn:blyth/np.git
Bash Function Tip : Define Bash function "t"
Useful bash function "t" shows definition of argument:
t(){ typeset -f $*; }
Example, apply "t" to itself:
epsilon:~ blyth$ t t
t ()
{
typeset -f $*;
: opticks.bash
}
The colon ":" line is a comment, here identifying where defined.
IPython Launch Customization via Bash Function "i"
epsilon:~ blyth$ t i
i ()
{
local opt="";
[ -n "$DEBUG" ] && opt="--debug";
${IPYTHON:-ipython} $opt --matplotlib --pdb -i $*;
: ~/.python_config;
: see also ~/.ipython/profile_default/ipython_config.py
}
epsilon:~ blyth$ DEBUG=1 i
[TerminalIPythonApp] IPYTHONDIR set to: /Users/blyth/.ipython
[TerminalIPythonApp] Using existing profile dir: '/Users/blyth/.ipython/profile_default'
[TerminalIPythonApp] Searching path ['/Users/blyth', '/Users/blyth/.ipython/profile_default', '/Users/blyth/miniconda3/etc/ipython', '/usr/local/etc/ipython', '/etc/ipython'] for config files
[TerminalIPythonApp] Attempting to load config file: ipython_config.py
[TerminalIPythonApp] Looking for ipython_config in /etc/ipython
[TerminalIPythonApp] Looking for ipython_config in /usr/local/etc/ipython
[TerminalIPythonApp] Looking for ipython_config in /Users/blyth/miniconda3/etc/ipython
[TerminalIPythonApp] Looking for ipython_config in /Users/blyth/.ipython/profile_default
[TerminalIPythonApp] Loaded config file: /Users/blyth/.ipython/profile_default/ipython_config.py
[TerminalIPythonApp] Looking for ipython_config in /Users/blyth
[TerminalIPythonApp] Enabling GUI event loop integration, eventloop=osx, matplotlib=MacOSX
Using matplotlib backend: MacOSX
[TerminalIPythonApp] Loading IPython extensions...
[TerminalIPythonApp] Loading IPython extension: storemagic
[TerminalIPythonApp] Running code from IPythonApp.exec_lines...
[TerminalIPythonApp] Running code in user namespace:
[TerminalIPythonApp] Running code in user namespace: import os, sys, logging
[TerminalIPythonApp] Running code in user namespace: log = logging.getLogger(__name__)
[TerminalIPythonApp] Running code in user namespace: import numpy as np
[TerminalIPythonApp] Running code in user namespace: import matplotlib.pyplot as plt
[TerminalIPythonApp] Running code in user namespace: from mpl_toolkits.mplot3d import Axes3D
[TerminalIPythonApp] Running code in user namespace: os.environ["TMP"] = os.path.expandvars("/tmp/$USER/opticks")
[TerminalIPythonApp] Running code in user namespace: sys.path.append(os.path.expanduser("~"))
[TerminalIPythonApp] Running code in user namespace: np.set_printoptions(suppress=True, precision=3, linewidth=200)
[TerminalIPythonApp] Running code in user namespace:
[TerminalIPythonApp] Starting IPythons mainloop...
In [1]:
Python Data Science Handbook
- https://jakevdp.github.io/PythonDataScienceHandbook/
- a good resource to get comfortable with python
All these packages, features, can be overwhelming
- dont be overwhelmed
- just need basic familiarity
- then can find whatever you need, when you need it
Objective is NOT to learn commands, Aims:
- get comfortable with IPython
- learn to play/discover functionality from various python packages
View HTML Locally : To Avoid Network Issues
Use browser to download ZIP of html pages from:
OR copy from IHEP to your laptop using scp:
scp lxslc7.ihep.ac.cn:~blyth/g/PythonDataScienceHandbook-gh-pages.zip .
Expand the zip archive of html pages:
mkdir PDSH cp PythonDataScienceHandbook-gh-pages.zip PDSH/ cd PDSH unzip -l PythonDataScienceHandbook-gh-pages.zip # check paths in the zip unzip PythonDataScienceHandbook-gh-pages.zip ln -s PythonDataScienceHandbook-gh-pages PythonDataScienceHandbook ln -s PythonDataScienceHandbook-gh-pages/theme python -m http.server ## start local web server open http://localhost:8000/PythonDataScienceHandbook/index.html
Exercise 3 : IPython copy/paste follow along
As I copy/paste handbook items into IPython...
- follow along in browser looking at handbook html
- copy/paste from your browser into your local IPython session
- Following slides are bitmapped, they are just a backup.
- Continue in your browser looking at Handbook html.
IPython : 01.01-help-and-documentation
http://localhost:8000/PythonDataScienceHandbook/01.01-help-and-documentation.html
https://jakevdp.github.io/PythonDataScienceHandbook/01.01-help-and-documentation.html
- IPython_help_?
- IPython_source_??
- IPython_tab
IPython_help_?
IPython_source_??
IPython_tab
IPython : 01.03-magic-commands
http://localhost:8000/PythonDataScienceHandbook/01.03-magic-commands.html
https://jakevdp.github.io/PythonDataScienceHandbook/01.03-magic-commands.html
- IPython_paste
- IPython_cpaste
- IPython_run
- IPython_timeit
IPython_paste
IPython_cpaste
IPython_run
IPython_timeit
IPython : 01.05-ipython-and-shell-commands
http://localhost:8000/PythonDataScienceHandbook/01.05-ipython-and-shell-commands.html
https://jakevdp.github.io/PythonDataScienceHandbook/01.05-ipython-and-shell-commands.html
- IPython_shell
IPython_shell
NumPy : 02.00-introduction-to-numpy
http://localhost:8000/PythonDataScienceHandbook/02.00-introduction-to-numpy.html
https://jakevdp.github.io/PythonDataScienceHandbook/02.00-introduction-to-numpy.html
- NumPy_np
NumPy_np
NumPy : 02.01-understanding-data-types
http://localhost:8000/PythonDataScienceHandbook/02.01-understanding-data-types.html
https://jakevdp.github.io/PythonDataScienceHandbook/02.01-understanding-data-types.html
- NumPy_array
- NumPy_zeros
- NumPy_types
NumPy_array
NumPy_zeros
NumPy_types
NumPy : 02.02-the-basics-of-numpy-arrays
http://localhost:8000/PythonDataScienceHandbook/02.02-the-basics-of-numpy-arrays.html
https://jakevdp.github.io/PythonDataScienceHandbook/02.02-the-basics-of-numpy-arrays.html
- NumPy_slice
- NumPy_subarray
- NumPy_reshape
NumPy_slice
NumPy_subarray
NumPy_reshape
NumPy : 02.03-computation-on-arrays-ufuncs
http://localhost:8000/PythonDataScienceHandbook/02.03-computation-on-arrays-ufuncs.html
https://jakevdp.github.io/PythonDataScienceHandbook/02.03-computation-on-arrays-ufuncs.html
- NumPy_wrong
- NumPy_right
- NumPy_scipy
NumPy_wrong
+----------------------+ | Large Loops in NumPy | | almost always | | wrong approach | | (very sloooooooow) | +----------------------+
NumPy_right
+----------------------+ | Correct approach | | MUCH MUCH FASTER | | ALSO SIMPLER | +----------------------+
NumPy_scipy
NumPy : 02.05-computation-on-arrays-broadcasting
http://localhost:8000/PythonDataScienceHandbook/02.05-computation-on-arrays-broadcasting.html
https://jakevdp.github.io/PythonDataScienceHandbook/02.05-computation-on-arrays-broadcasting.html
- NumPy_broadcasting
NumPy_broadcasting
NumPy : 02.06-boolean-arrays-and-masks
http://localhost:8000/PythonDataScienceHandbook/02.06-boolean-arrays-and-masks.html
https://jakevdp.github.io/PythonDataScienceHandbook/02.06-boolean-arrays-and-masks.html
- NumPy_mask
- NumPy_count_nonzero
- NumPy_masking
NumPy_mask
NumPy_count_nonzero
NumPy_masking
Matplotlib : 04.00-introduction-to-matplotlib
http://localhost:8000/PythonDataScienceHandbook/04.00-introduction-to-matplotlib.html
https://jakevdp.github.io/PythonDataScienceHandbook/04.00-introduction-to-matplotlib.html
- Matplotlib_script
- Matplotlib_IPython
- Matplotlib_IPython_demo
- Matplotlib_scatter
- Matplotlib_scatter_demo
Matplotlib_script
Matplotlib_IPython
Matplotlib_IPython_demo
Matplotlib_scatter
Matplotlib_scatter_demo
FewPMT_demo.png
Example of Matplotlib scatter plot, with some dotted lines
Python Data Science Handbook1
- https://jakevdp.github.io/PythonDataScienceHandbook/
- a good resource to get comfortable with python
We only skimmed the fundamental sections from book
Many more sections for you to explore, eg:
|
Now that you are familiar with IPython shell:
- you can easily continue exploring ...
sklearn_Logo
Three Laws of Good Software Design + Development
KEY : Units + Unit testing
Complex problems : Many simple units
- each unit with test ready to run
- dive into code in seconds
Tame complexity : Divide and Conquer
|
|
|
Lao Tzu
You need to learn so many things... : Actually no, you do not
- you need to learn (or re-learn) what you need, when you need it
Retain core principals that enable you to learn quickly
- unit tests : not just for testing
- hugely useful for learning+reminding too